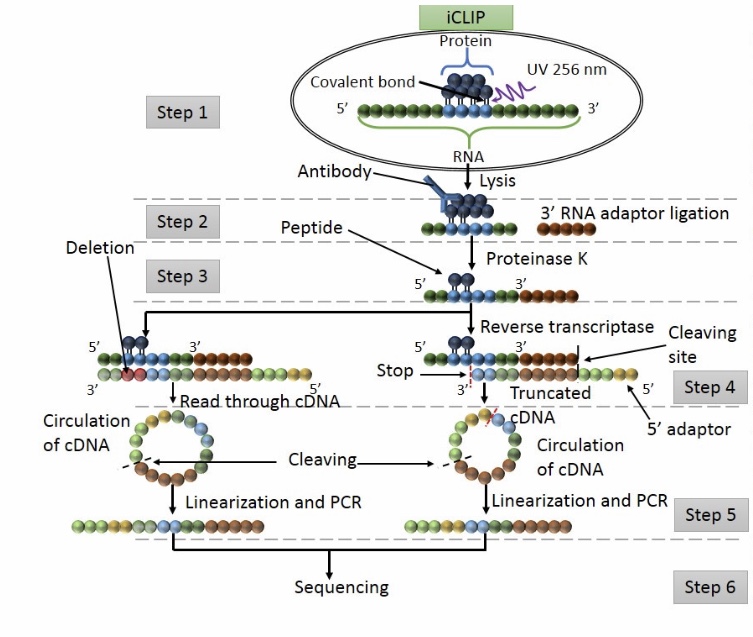
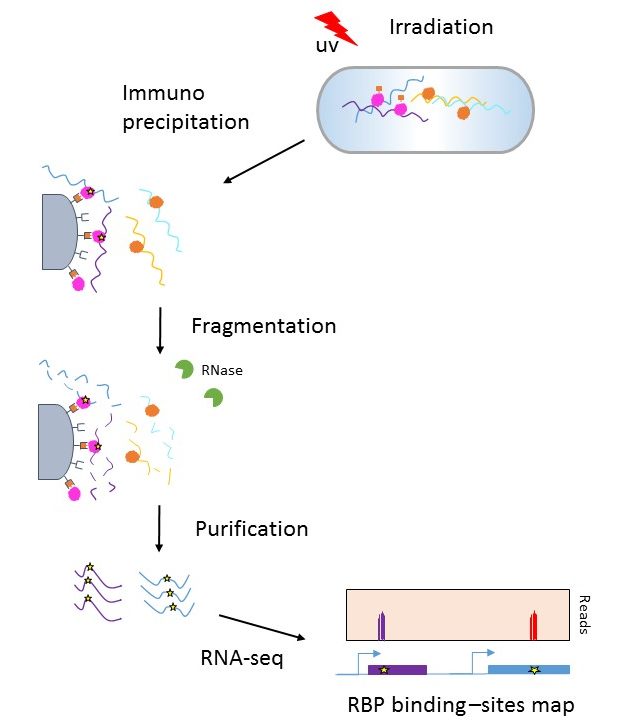
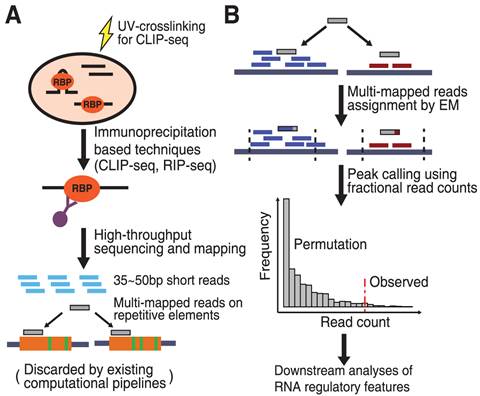
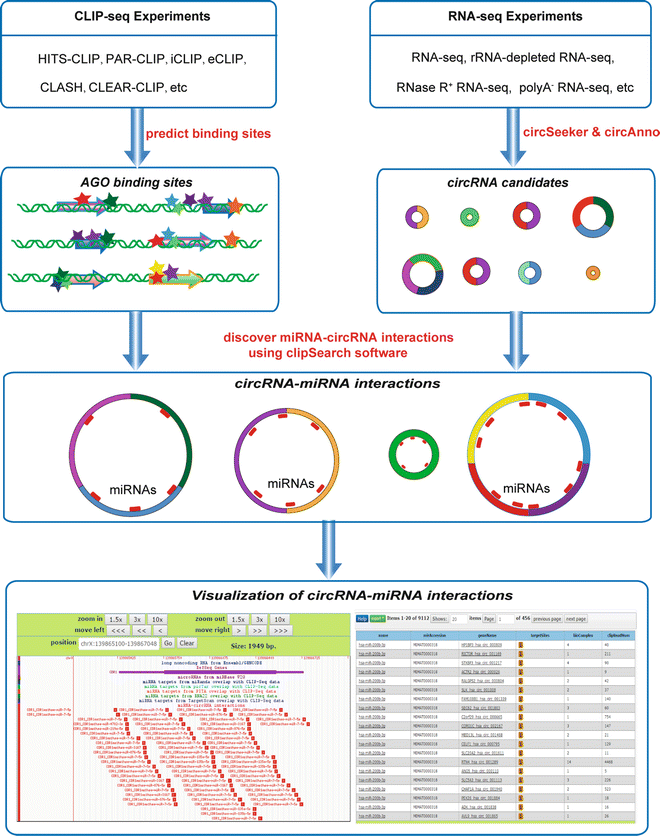
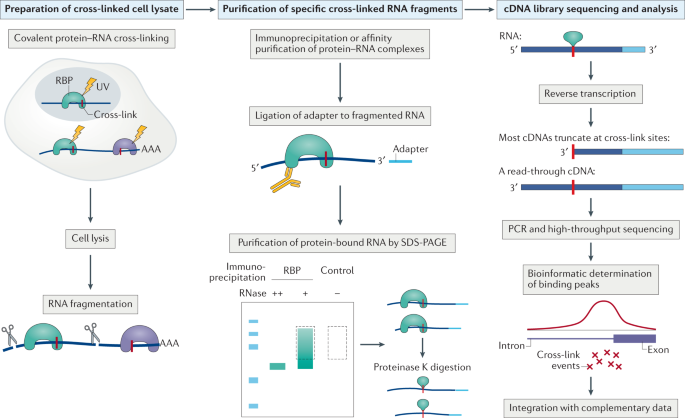
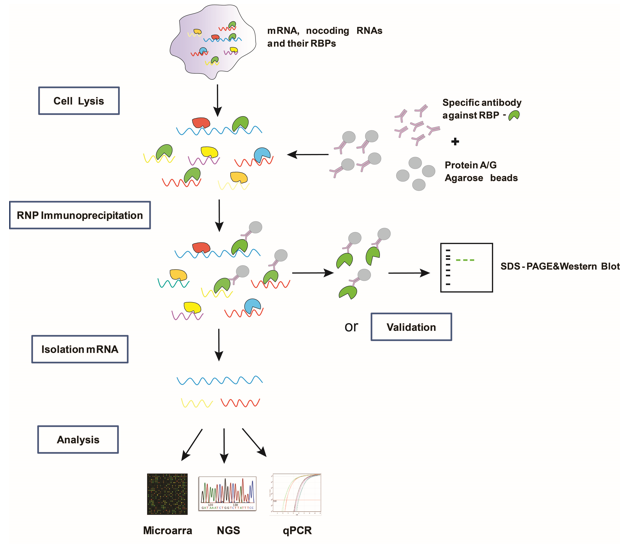
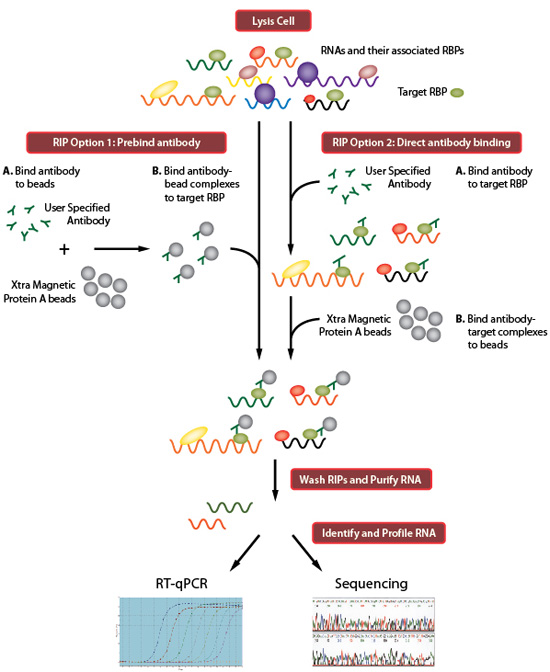
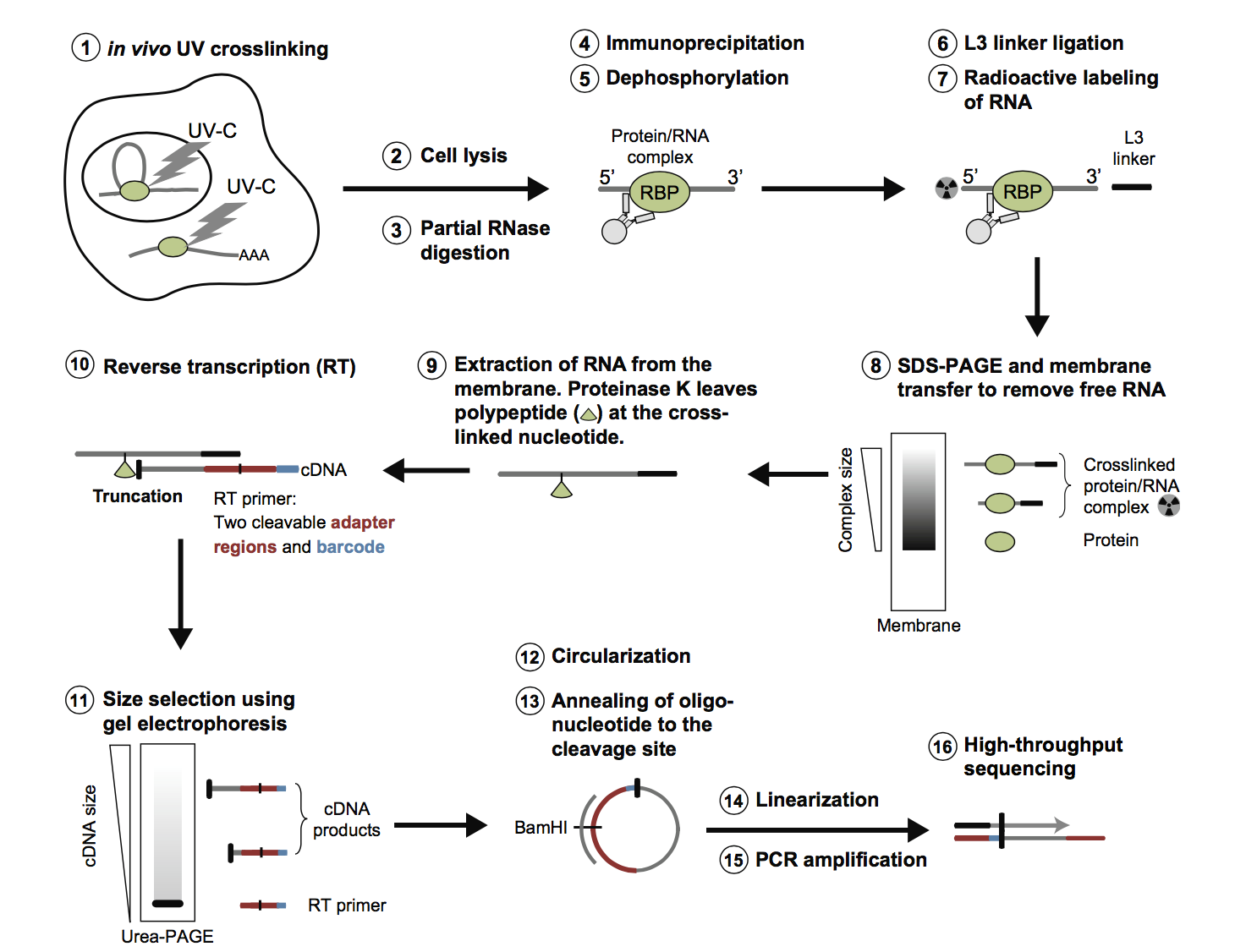
Genome-wide Mapping of Cellular Protein–RNA Interactions Enabled by Chemical Crosslinking - ScienceDirect

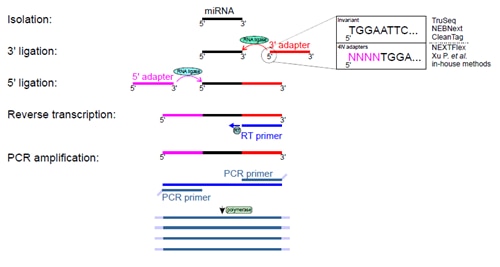
PAR-CLIP and Streamlined Small RNA cDNA Library Preparation Protocol for the Identification of RNA Binding Protein Target Sites | RNA-Seq Blog

Enhanced Crosslinking Immunoprecipitation (eCLIP) Method for Efficient Identification of Protein-bound RNA in Mouse Testis | Protocol
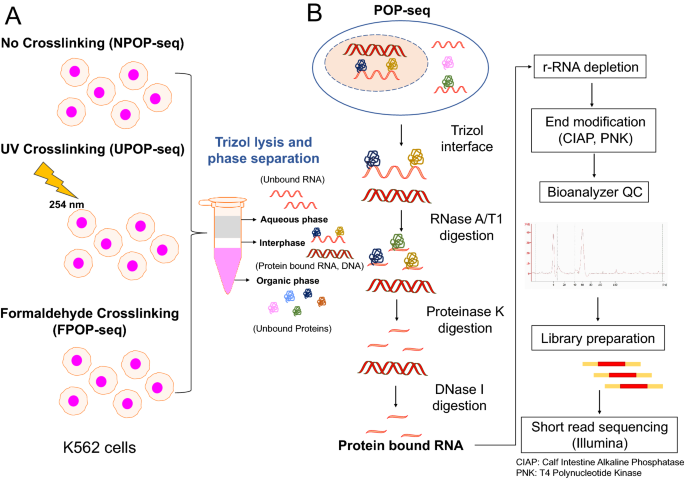
Transcriptome-wide high-throughput mapping of protein–RNA occupancy profiles using POP-seq | Scientific Reports

Pipeline for extracting MIWI CLIP-Seq features (see Section 2). The RNA... | Download Scientific Diagram
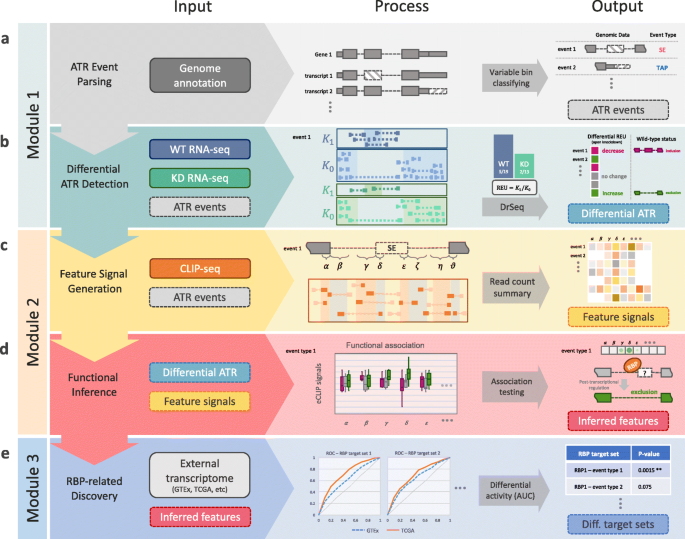
SURF: integrative analysis of a compendium of RNA-seq and CLIP-seq datasets highlights complex governing of alternative transcriptional regulation by RNA-binding proteins | Genome Biology | Full Text

Flow diagram of CLIP-seq. Cells are fed with the ribonucleoside analog... | Download Scientific Diagram